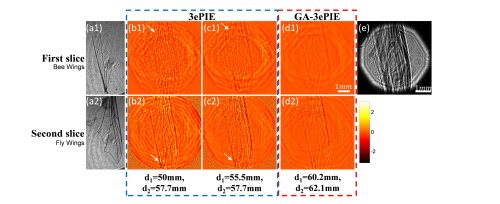
A multiple slices approach in ptychography, termed 3ePIE algorithm, solves the multiple scattering problems of thick samples, which is crucial to biomedical imaging and in situ studies. However, it is unclear how many slices need to be separated and what the distance is between each layer respectively, while these two parameters are sensitive and crucial to the recovery of thick samples. The traditional method is to separate the sample with the same interval for convenience and to test the number of sections for the best reconstructions, which is reasonable for continuous samples and has achieved great results. But this kind of segmentation approach may not be scientific enough for those discrete samples with an uneven spatial distribution. The two inaccurate parameters may yield the algorithm to diverge or generate artifacts, and the empty slices may decrease the reconstruction quality and increase computation time. In addition, repeatedly testing the number of slices is tedious work even for a continuous sample. To this end, Baoli Yao’s research team proposed a genetic algorithm-based 3ePIE approach, termed the GA-3ePIE method, to retrieve both the interval between each layer and the number of slices. The performance is verified by both simulations and experiments. The maximum number of sections that can be resolved is also investigated in numerical analysis, which is associated with the sampling and overlap rate in a spatial domain. Their method can be also promoted to image thick samples with coherent X-rays and in the electron regime. The limitations of our method are also discussed.
(Original research article "Optics Express Vol. 27, Issue 4, pp. 5433-5446 (2019) https://www.osapublishing.org/oe/abstract.cfm?uri=oe-27-11-15693")
Download: