Facultative intracellular pathogens are able to live inside and outside host cells. It is highly desirable to differentiate their cellular locations for the purposes of fundamental research and clinical applications. In this work, we developed a novel analysis platform that allows users to choose two analysis models: amplitude weighted lifetime (τA) and intensity weighted lifetime (τI) for fluorescence lifetime imaging microscopy (FLIM). We applied these two models to analyse FLIM images of mouse Raw macrophage cells that were infected with bacteria Shigella Sonnei, adherent and invasive E. coli (AIEC) and Lactobacillus. The results show that the fluorescence lifetimes of bacteria depend on their cellular locations. The τA model is superior in visually differentiating bacteria that are in extra- and intra-cellular and membrane-bounded locations, whereas the τI model show excellent precision. Both models show speedy performances that analysis can be performed within 0.3 s. We also compared the proposed models with a widely used commercial software tool (τC, SPC Image, Becker & Hickl GmbH), showing similar τI and τC results. The platform also allows users to perform phasor analysis with great flexibility to pinpoint the regions of interest from lifetime images as well as phasor plots. This platform holds the disruptive potential of replacing z-stack imaging for identifying intracellular bacteria.
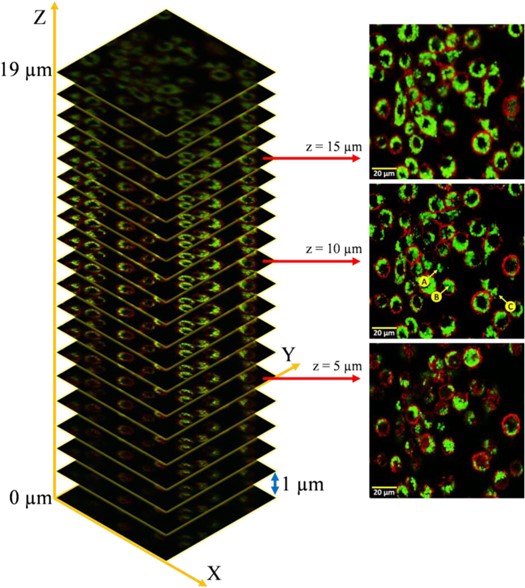
z-stack images of Shi86 and macrophage cells at (a) upper, (b) middle, and (c) lower levels. (Image by XIOPM)
Download: