DNA metabolism is an up to date subject being studied worldwide. The inner mechanism is complicated, of which researchers are devoted to observe more precisely and distinctly.
During DNA metabolism, a specific protein named single-stranded DNA binding protein (SSB) plays key roles in facilitating the repair processes of DNA damage. However, visualizing SSB localization and ascertaining potential spatiotemporal changes in response to DNA damage is difficult to observe.
In order to distinguish the process out of background environment contexts, a lot of research efforts have been done, and the phenomena is been observed by super-resolution imaging recently.
A research team led by Prof. Dr. Ming Lei from Xi'an Institute of Optics and Precision Mechanics (XIOPM) of the Chinese Academy of Sciences (CAS) developed a novel, super-resolution microscope optimized for the study of prokaryotic cells, which helps observing the whole process of its inner mechanism. The result was published in Genes to Cells.
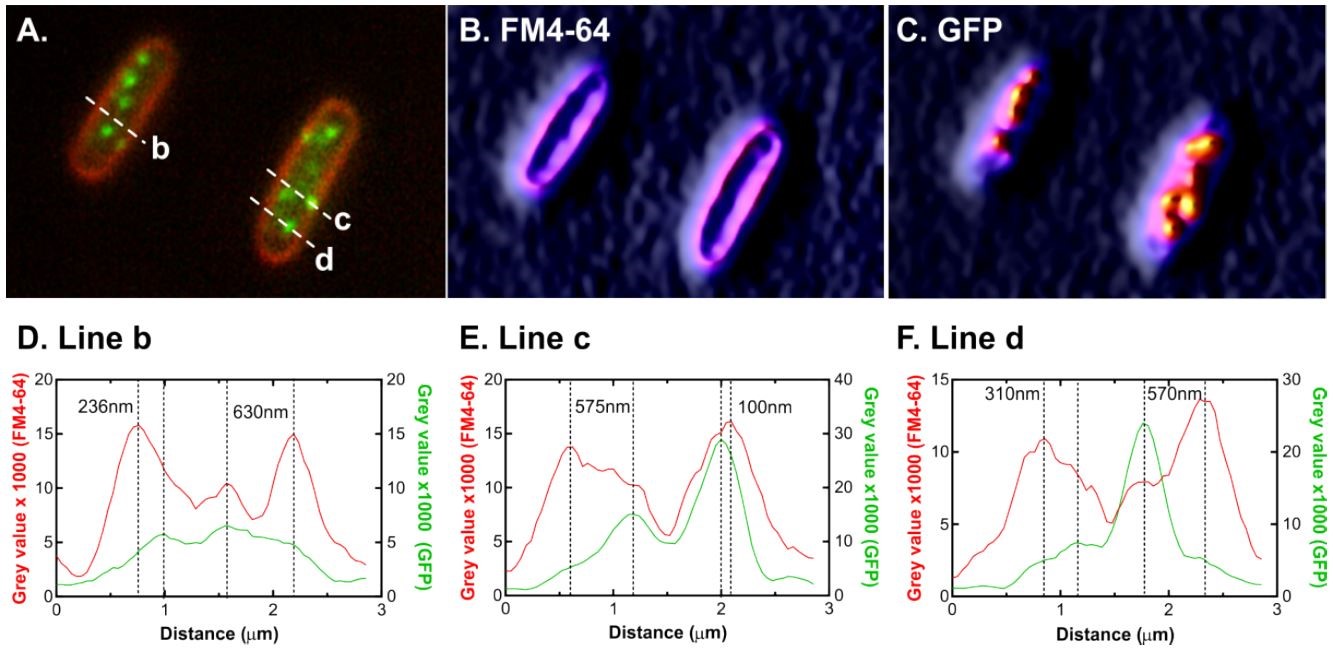 SSB is no longer associated with the inner membrane following DNA damage (Image by XIOPM)
SSB is no longer associated with the inner membrane following DNA damage (Image by XIOPM)
The super-resolution imaging system comprised of a synchronized, structured illumination microscope cooperated with an image reconstruction algorithm. It is a novel super-resolution microscope especially designed for low light imaging. Meanwhile, it is proved ideal for clear discernment of structures within observation cells.
The structures been observed among cells is SSB, the E. coli SSB is essential to viability. It plays key roles in DNA metabolism where it binds to nascent single strands of DNA and to target proteins known as the SSB interactome.
Observing results show that SSB normally localizes to a small number of foci and the excess protein is associated with the inner membrane. However, when DNA damage occurs, SSB disengages from the membrane and move into cells, spreading over the genome and enabling repair events. Once the damage been repaired, SSB returns back to the inner membrane.
These findings suggest that a control mechanism for maintaining SSB at the inner membrane utilizing phospholipid/OB-fold interactions and a means for SSB to rapidly deliver repair enzymes to the genome when their actions are required, which revealed the inner mechanism of DNA metabolism. Furthermore, more observation wound be done on DNA replication to fully reveal the DNA metabolic mechanism among cells.
Download: