Fluorescence nanoscopy has become an indispensable tool for studying organelle structures, protein dynamics, and interactions in biological sciences. Single-molecule localization microscopy can now routinely achieve 10–50 nm resolution through fluorescently labeled specimens in lateral optical sections.
However, visualizing structures organized along the axial direction demands scanning and imaging each of the lateral imaging planes with fine intervals throughout the whole cell. This iterative process suffers from photo bleaching of tagged probes, is susceptible to alignment artifacts and also limits the imaging speed.
Here, we focused on the axial plane super-resolution imaging which integrated the single-objective light-sheet illumination and axial plane optical imaging with single-molecule localization technique to resolve nanoscale cellular architectures along the axial (or depth) dimension without scanning.
A doctor candicate—AN Sha from Xi'an Institute of Optics and Precision Mechanics (XIOPM) of the Chinese Academy of Sciences (CAS) demonstrated that this method is compatible with DNA points accumulation for imaging in nanoscale topography (DNA-PAINT) and exchange-PAINT by virtue of its light-sheet illumination, allowing multiplexed super-resolution imaging throughout the depth of whole cells.
They further demonstrated this proposed system by resolving the axial distributions of intracellular organelles such as microtubules, mitochondria, and nuclear pore complexes in both COS-7 cells and glioblastoma patient-derived tumor cells.
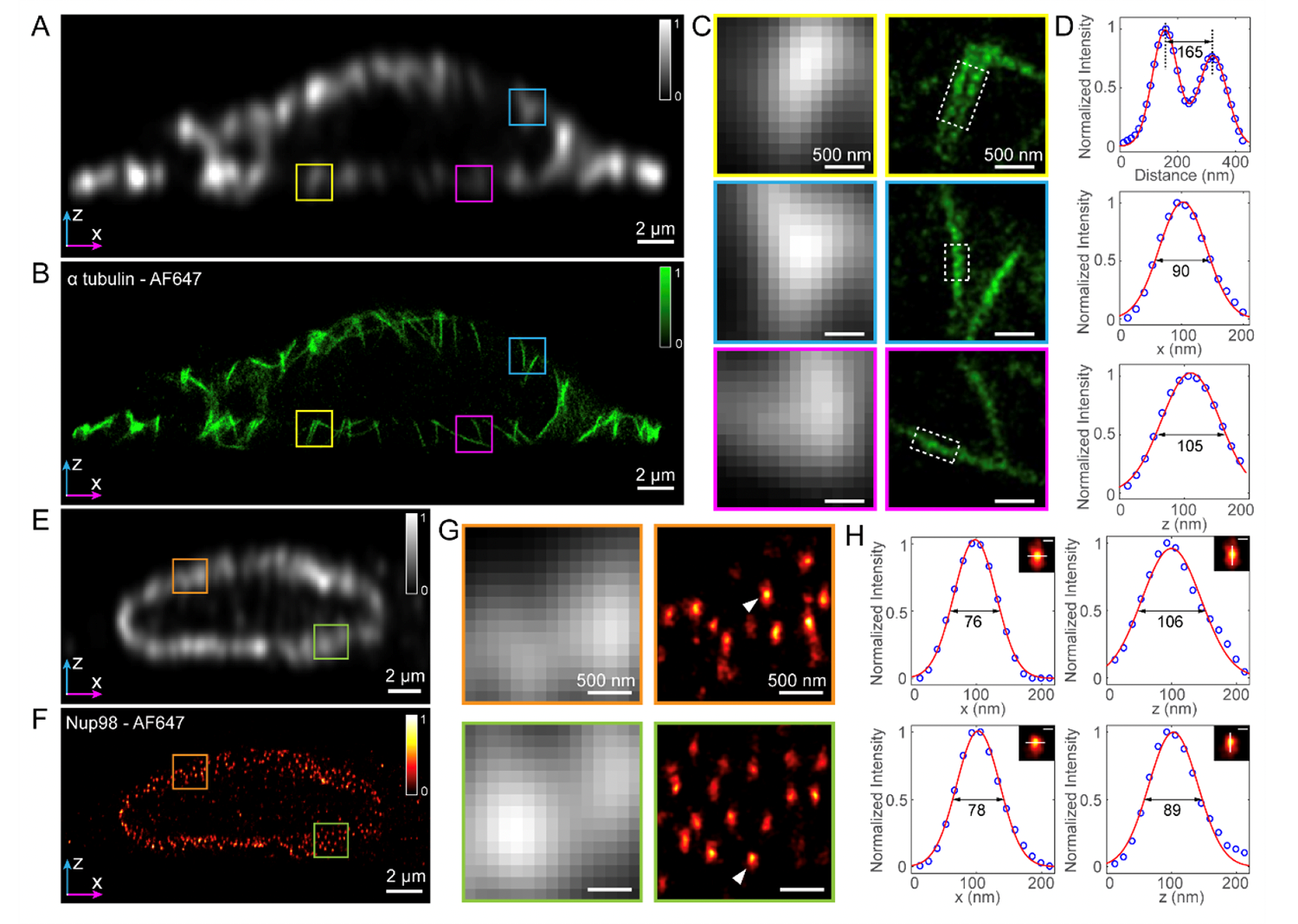 Super-resolution reconstructions of microtubules and nuclear pore complexes by the axial plane SMLM in fixed COS-7 cells. (Image by XIOPM)
Super-resolution reconstructions of microtubules and nuclear pore complexes by the axial plane SMLM in fixed COS-7 cells. (Image by XIOPM)
(Original research article “Biomedical Optics Express” (2020) https://doi.org/10.1364/BOE.377890 )


